The on-going pandemic spread of Severe Acute Respiratory Syndrome Coronavirus -2 (SARS-CoV-2) requires fast and easy to implement detection methods. The virus, being the causative agent of COVID-19 disease is most frequently detected by RT-qPCR assay. This assay combines a very high sensitivity and specificity characteristic for the nucleic acid detection assays. During the assay single stranded RNA genome is converted to DNA which is detected by qPCR (also known as Real-Time PCR). Currently a lot of suppliers are offering validated assays for the detection of SARS-CoV-2 genome by RT-qPCR. In spite of the wide adoption of this technology, at present the limiting step in achieving fast and easy to implement SARS-CoV-2 detection is the sample preparation step. The detection step by RT-qPCR can take somewhere between 45 minutes and 2 hours and 96 to 384 samples in parallel can be analysed per single qPCR instrument. In contrast, the sample preparation at least doubles this time taking between 60 minutes and 4 hours to prepare this number of samples for analysis. Usually the samples are prepared by RNA extraction using column based or magnetic beads nucleic acid absorption. This is not only time and resource consuming, but also leads to the loss of nucleic acids thus reducing the sensitivity. Both, time and costs are of special interest in pandemic environment where important shortage of materials is to be expected.
Here we describe an extremely simple 5 minutes sample preparation step and its successful application for the detection of SARS-CoV-2 (Fig.1).
The samples were prepared as described in Swab DNA/ RNA isolation application note. Shortly, the sample was collected with viscose swab and extracted by submerging immediately the sampling end in 200 μl of DireCtQuant 100W. The samples were then heated at 90ºC with agitation. 1μl of the sample was subsequently used to prepare 20 μl one-step RT-qPCR reaction using the recommended protocol (1) The detection was performed by multiplex detection using CFX96Touch instrument (Bio-Rad).
The SARS-CoV-2 detection was performed by amplification of envelope (E) gene and RNA-dependent RNA Polymerase (RdRp) gene as recommended by World Health Organization (1). Internal control of human RNA sequence was used as a positive control for sample validity, this proves correct sample preparation and processing and avoids false negative results. The probes used for detection ware labelled with FAM (E gene), HEX (RdRp gene) and Cy5 (human RNA sequence).
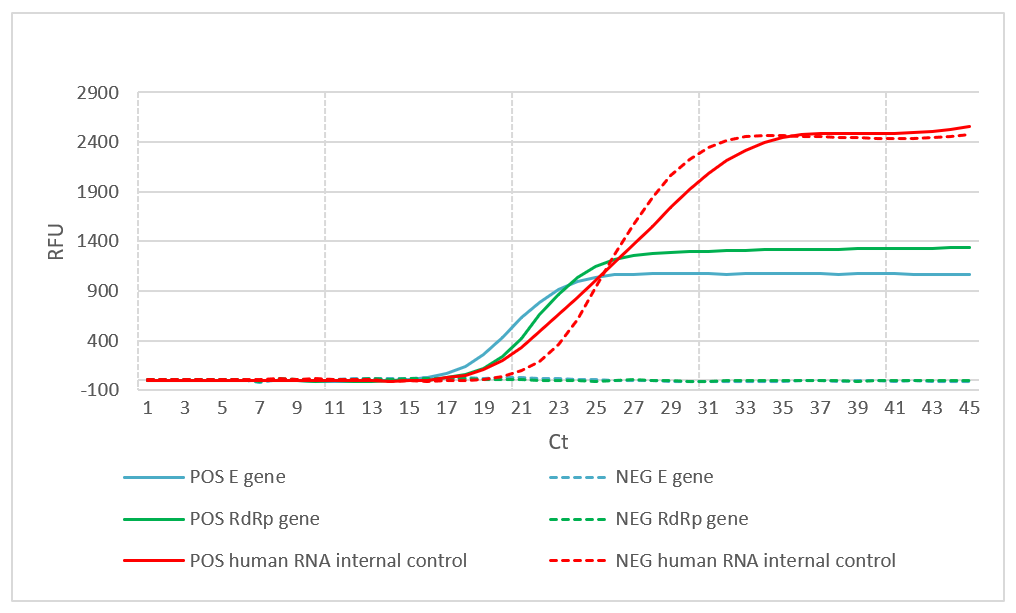
Figure 1. Detection of SARS-CoV-2 using fast DireCtQuant workflow. A swab sample was prepared according to Swab DNA/ RNA isolation application note . The detection was performed using 1 μl of sample in 20 μl One-Step RT-qPCR reaction. Solid lines represent SARS-CoV-2 positive sample (POS), dotted lines represent SARS-CoV-2 negative sample (NEG). Note the confident detection of the internal control in both NEG and POS samples.
In conclusions, using the DireCtQuant sample preparation the total detection time is reduced from 3 hours (1h 30min sample preparation plus 1h 30min RT-qPCR) to 1 hour 40 minutes or 45% reduction in the required time (Fig.2).
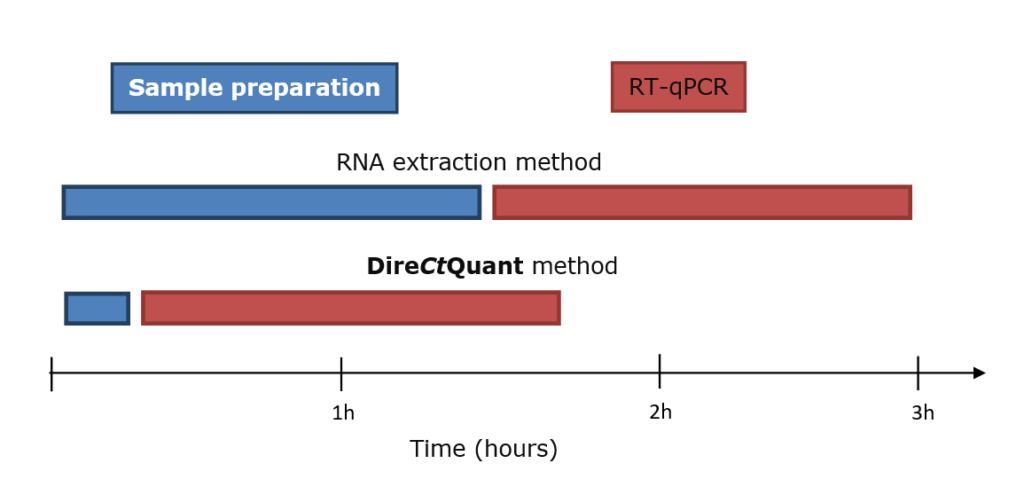
Figure 2. Time comparison between RNA extraction method and DireCtQuant method for detection of SARS-CoV-2
The simplicity and the heat extraction step reduce significantly the biohazard risk associated with testing infectious material. A huge decrease of the required work, consumables and cost are achieved. The cost per reaction (approximately 2 euros) is several times less than the majority of the column or magnetic beads-based approaches. The prepared sample can be stored for long periods of time without degradation for the confirmatory and archival purposes. This is achieved by the unique DireCtQuant formulation including Stop-the-TimeTM technology. The protocol could be extended to different types of starting material such as saliva samples, surface sample, etc. Another advantage of the method presented here is the increase in the sensitivity compared to RNA extraction method as here no extraction is performed and all the RNA present in the sample can be used for detection without any loss.
References:
- Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Corman et al. Euro Surveill. 2020 Jan;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045.