This protocol is designed and validated for quantitative extraction of bacteria, viruses (DNA and RNA) and epithelial cells collected with swabs in less than 5 minutes using a single tube. If the aim of the study is bacterial or virus DNA/RNA detection/quantification genomic DNA from epithelial cells within the sample can be used as positive control for correct sample collection and extraction.
Additional equipment:
- Snap tool or scissors
- Thermoblock (with agitation is preferred) or cup of boiling water
- Centrifuge reaching 10.000g. (If the centrifuge is reaching only 3.000g the centrifugation time should be increased as indicated below).
The protocol is identical for DireCtQuant 100ST (#DCQ100ST) and DireCtQuant 100W (#DCQ100W).
For illustration purposes #DCQ100ST is used.
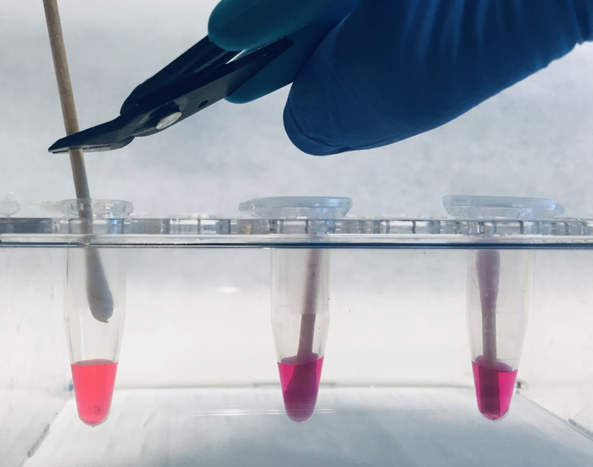
- Pipet 200 μl of #DCQ100ST or #DCQ100W in a 1.5 ml tube.
- Snip the swab applicator as high as possible, permitting the tube to be closed.
- Close the tube. Make sure that the swab is completely wetted by the reagent.
- Incubate 3 minutes at 90°C. If possible, apply agitation during the incubation.
- Leave the tube 1 minute on the bench to cool down.
- With a sterile tool reverse the swab upside down.
- Close the tube and centrifuge at 10.000g for 30s in order to collect all the liquid absorbed in the swab. For Minicentrifuge reaching only 3.000g increase time to 3 minutes.
- Discard the swab.
- The samples can be stored and transported at room temperature after the extraction. For more information about the lon-term storage conditions check DireCtQuant 100ST (#DCQ100ST) or DireCtQuant 100W (#DCQ100W) protocols respectively.
- Use 1.0 μl of the sample per 20.0 μl PCR reaction. The extracted sample should be enough for around 200 detections. For more details about the PCR reaction conditions check DireCtQuant 100ST (#DCQ100ST) or DireCtQuant 100W (#DCQ100W) protocols respectively.